ELECTROANALYTICAL BIOSENSING PLATFORMS FOR THE DETERMINATION OF METHYLATIONS IN NUCLEIC ACIDS AT GLOBAL AND REGIONAL LEVELS
|
Description |
The developed technologies allow the individual or simultaneous determination of the main methylated bases in DNA (5-methylcytosine, 5-mC and 5-hydroxymethylcytosine, 5-hmC) and RNA (N6-methyladenosine, m6A) both globally and regionally in a fast, reliable and simple way. The developed methodologies are based on the use of amperometric biosensors that employ micromagnetic carriers (MBs) functionalized with suitable bioreceptors and amperometric detection on disposable screen-printed electrodes.
In these biodevices the amperometric signal obtained, which can be related to the amount of target methylated base, is based on the enzymatic and electrochemical reactions involved in the horseradish peroxidase (HRP)/hydroquinone (HQ)/H2O2 system that take place after the capture of the MBs on the surface of a disposable electrochemical transducer at an appropriate constant potential.
|
How does it work |
The technologies are based on the use of MBs modified with a synthetic DNA sequence complementary to the region of the gene where methylation is to be detected, for determination at the regional level (Figure 1a), or with commercial antibodies selective to the methylated base to be determined, for determination at the global level (Figure 1b).
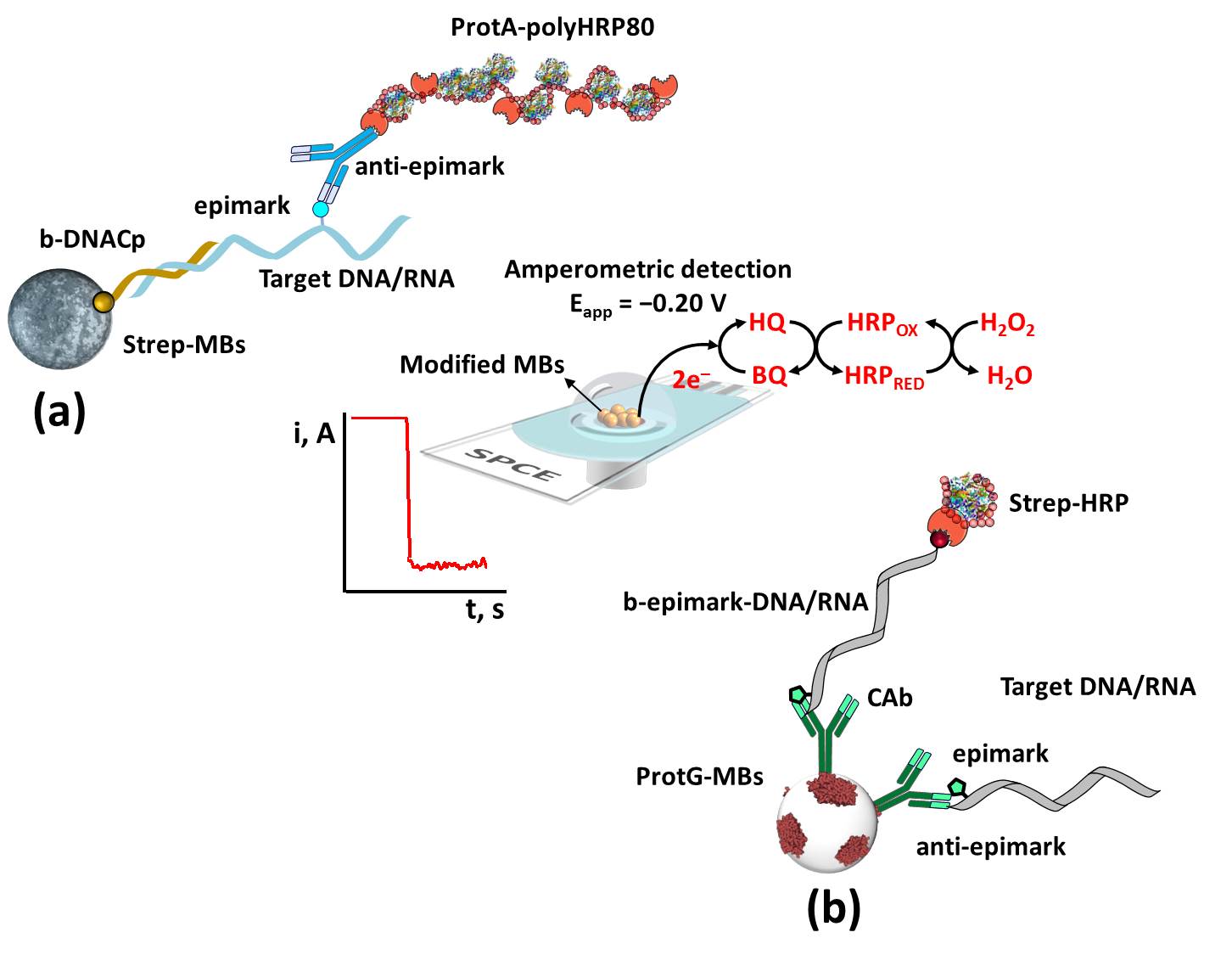
Figure 1. Schematic diagram of the biosensing platforms developed for the determination of methylations in nucleic acids at regional (a) and global (b) levels.
For determination at the regional level (Figure 1a), the methylated target DNA or RNA is selectively captured on modified MBs with a synthetic biotinylated DNA probe that hybridizes to the region adjacent to that where the presence of methylations is to be assessed. The methylated nucleic acid is then detected using an antibody selective to the target methylated base that is enzymatically labeled with HRP using commercially available bioreceptors with multiple HRP molecules (Protein A-polyHRP80).
For global determination (Figure 1b), direct competitive immunoassay formats are performed on magnetic immunoconjugates in which methylated target DNA or RNA competes for the limited binding sites of the antibodies attached to the MBs with a synthetic biotinylated DNA or RNA oligomer carrying a single methylated base that is labeled with HRP enzyme using the appropriate commercial reagent (streptavidin-HRP conjugate).
In all cases, the modified MBs are magnetically captured on the surface of disposable electrodes (or electrode arrays) and amperometric transduction is performed using the HRP/ HQ/H2O2 system yielding a cathodic current variation attributed to the HQ-mediated enzymatic reduction of H2O2, which can be related to the concentration of the target methylated base.
|
Advantages |
Among the competitive advantages of the developed methodologies are the following:
- Easily transferable to the determination of other epimarks of interest.
- Due to the type of detection and instrumentation used, simple implementation in point-of-care (POC) or "sample-result" diagnostic devices.
- Possibility of simultaneous determination of 5-mC and 5-hmC, the two most frequent and similar methylated bases in DNA.
- Sensitivity at the level of a single base without the need to use nucleic acid amplification steps.
- Capable of operating at room temperature.
- Provide quantitative results in less than 2 h and at a very affordable cost (< 2 €/determination).
|
Where has it been developed |
These bioplatforms have been developed at the Electroanalysis and Electrochemical (Bio)Sensors Group of the Faculty of Chemical Sciences of the Complutense University of Madrid (GEBE Group, UCM Group Reference 910319) and in collaboration with researchers and clinicians from the Carlos III Health Institute and the La Paz University Hospital.
|
And also |
The results obtained confirm excellent performance for reliable determinations in cells, human biopsies of different nature (solid and liquid) and directly in human serum without prior extraction of genomic DNA.
The potential of these bioplatforms to discriminate between healthy and cancer-affected individuals has also been demonstrated by determining the overall level of 5-mC and 5-hmC in genomic DNA extracted from tissues (sensitivity and specificity of 89 % and 67 %, respectively) or by detecting 5-mC at the regional level directly in serum without prior extraction of genetic material. In addition, they allow characterization of the cancer cells aggressiveness of by detecting m6A at the global level in a small amount of total RNA extracted without the need for enrichment or fragmentation. In determinations in human cells and tissues, the amount of genomic DNA or cellular RNA used per determination is between 10 and 100 ng.
The incorporation of these biotools, which deliver results consistent with those provided by conventional methodologies (RT-PCR), into routine clinical analysis, would entail important advantages in terms of simplicity, speed, cost, portability and possibility of multiplexing, reliability for the early diagnosis and prognosis of prevalent diseases such as cancer and improvement of the efficiency of the applied therapy.
Website with additional information: https://gebeucm.wordpress.com/
|
Contact |
|
© Office for the Transfer of Research Results – UCM |
|
PDF Downloads |
|
Classification |
|
Responsible Researchers |
Susana Campuzano Ruiz: susanacr@quim.ucm.es
María Pedrero Muñoz: mpedrero@quim.ucm.es
José M. Pingarrón Carrazón: pingarro@quim.ucm.es
Department: Analytic chemistry
Faculty: Chemical Sciences